Empty Pipes
The 20 Most Studied Genes
The Nation Center for Biotechnology Information (NCBI) maintains an enormous amount of biological data and provides it all to the public for no cost as a collection of databases. One of the most popular is GenBank, which contains information about annotated genes. Consider the gene p53, which encodes a tumor suppressor protein, the absence of which allows many cancers to proliferate. By looking at its entry in GenBank, we can immediately find out its full name (tumor protein p53), which organism this entry corresponds to (Human), aliases (BCC7, LFS1, TRP53), a short description and a whole host of other technical information.
Among the information provided with each entry is a section which contains a list of papers which have referenced this gene. In a sense, each reference is a paper which has contributed some bit of knowledge about the function of this piece of DNA (or RNA). This got me wondering, which are the most studied genes? Which genes have made an appearance in the most published papers?
To answer this, I downloaded the table which contains the reference information from GenBank, performed some rudimentary analysis, and generated the following table of the top 20 most popular genes, as measured by the number of times they have been cited:
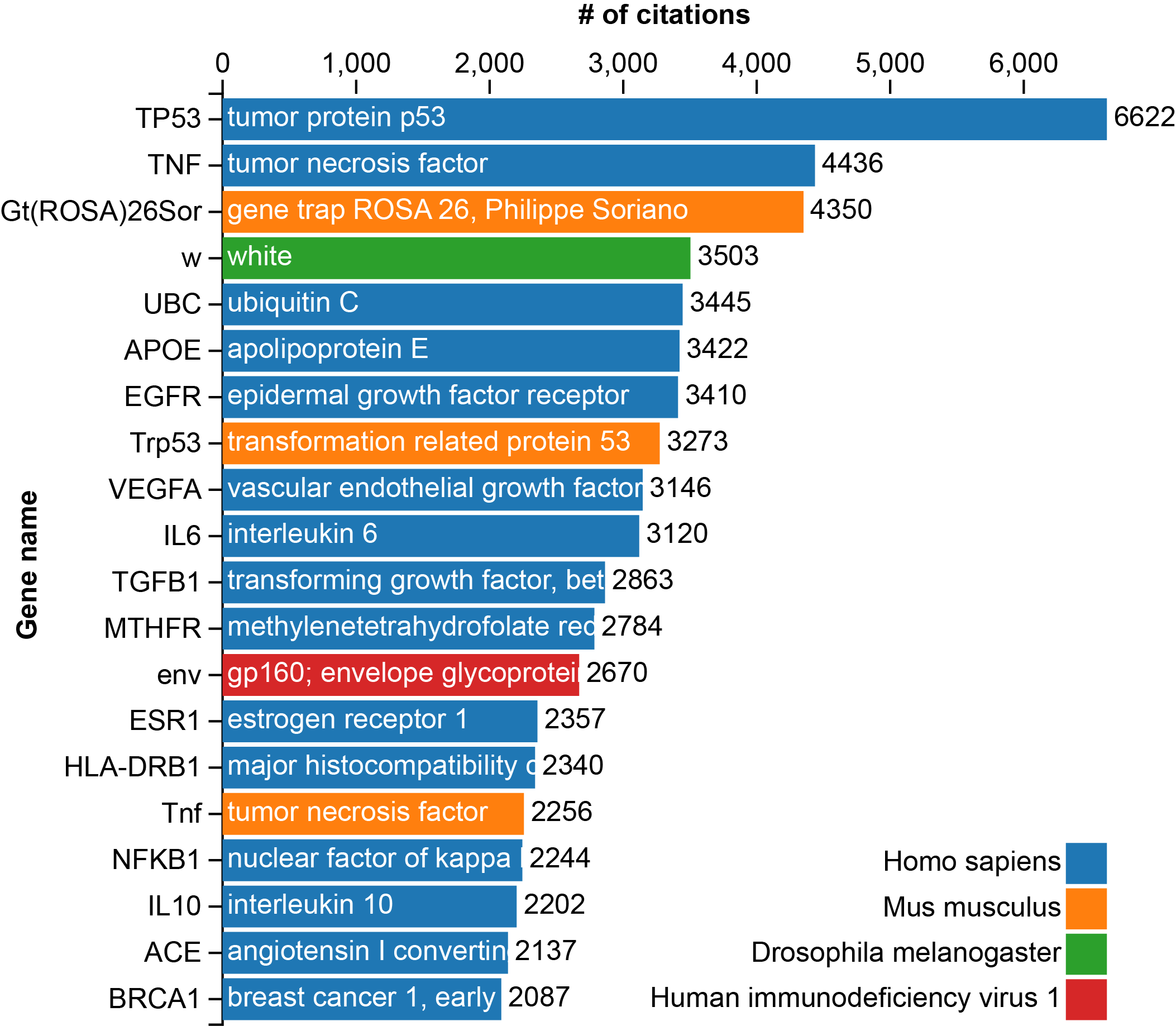
The graph above shows the number of references in PubMed to a particular gene in GenBank. The color of the bars refers to the organism that the gene is found in. It was made using d3.js and the script for generating it can be found here (github.com), while the data itself is located here (github.com).
The genes on the list can be broadly placed into 6 categories:
-
Cancer related - Tp53, Trp53 and all of the genes with ‘cancer’ (BRCA1), ‘tumor’ (TNF) or ‘growth factor’ (EGFR, VEGFA and TGFB) in their name are likely associated with cancer and are involved in either helping cells proliferate (oncogenes) or preventing them from becoming cancerous (tumor suppressor genes).
-
Immune system related - Interleukins, ‘nuclear factor kappa-light-chain-enhancer of activated B cells’ (also known as NF-κB) and major histocompatibility complex (MHC) are all associated with immune responses such as recognizing pathogens and mounting an attack against them.
-
HIV related - gp160 envelope glycoprotein (env) is one of the proteins on the surface of retroviruses which allow it to attach to and enter cells. Needless to say, it is extremely important in finding treatments and vaccines for such viruses.
-
Other disease - Apolipoprotein E (APOE) is involved in heart disease and Alzheimer’s disease, while Methylenetetrahydrofolate reductase (MTHFR) is associated with susceptibility to a variety of disorders including Alzheimer’s, colon cancer and others.
-
Regulatory - Ubiquitin (UBC) is a protein involved in the translocation and degradation (among other processes) of other proteins. Angiotensin-converting enzyme (ACE) is a regulatory enzyme which is involved in the control of blood pressure. Estrogen receptor 1 (ESR1) is a transcription factor which responds to the hormone estrogen, leading to a variety of downstream effects.
-
Other - The gene w (white), is popular largely due to its historical cachet. It was the first mutation to be discovered which did not display typical Mendelian inheritance due to its location on a sex-chromosome in D. melanogaster. Gene trap ROSA 26 (gt(ROSA)26Sor) is simply a convenient place to insert genes for study in a mouse model.
Immediately evident is the overrepresentation of disease-related genes. 15 of the 20 genes are heavily involved in some human disease. The remaining entries are either regulatory (UBC, ACE and ESR1), historic (w) or just simply useful (Gt(ROSA)26Sor). The majority come from human, followed by mouse (used to express genes also found in humans: Tnf and Trp53), and finally HIV and Drosophila. This is something of a reflection of where our interests and funding lie. The two most studied genes are involved in cancer, research in which is both well-funded and heavily reliant on genetic analysis. Four on the list are associated with the immune system (IL6, IL10, NFKB1, and HLA-DRB1), two (APOE and ACE) are associated with heart disease and one with HIV. We focus the majority of our attention on the things which are likely to kill us.
Conspicuously absent from the list are any genes from plants or genes involved in metabolism. Important pathways such as differentiation, DNA replication and protein synthesis are all absent. That’s not to say that they are not studied, it’s just that they recieve less attention than processes involved in our demise. Then again, the age of molecular genetics has only begun in the last century or so. Perhaps our interests will shift in the future as we find cures and treatments for existing maladies and start having to deal with others such as a a changing climate, energy crises and an aging population. Biology may hold partial solutions to these problems and the proportional amount of effort we put into finding processes to remove carbon dioxide from the air, to produce fuels from biomatter or to limit or reverse aging may grow to eclipse that put into research in the current top-20 genes.
Reviving a Seemingly Dead Tmux
Quite often, when I try to attach to an existing tmux session, the following error pops up:
failed to connect to server: Connection refusedIt seems like tmux has disappeared or crashed. Fortunately, to this date that
has never been the case. It’s just a simple case of a deleted socket. To cut a
long story short, fixing it requires sending tmux a signal to recreate the
socket:
killall -s SIGUSR1 tmuxHere’s a reference to a stackexchange question which gives slightly more information.
VIM Python Snippets
There are many times when a task calls for a simple python script. It is usually something small that takes some input file as a parameter, does some processing, and then spits out some results. It might even take an options or two. It’s tempting to just throw some lines of code into a file and be done with it. This may work but often just makes things more difficult later.
Consider the following code (let’s creatively call it do_stuff.py)
which simply converts the input to uppercase:
import sys
for line in sys.stdin:
print line.upper()What happens, however, when it grows a little bit and we add a function?
import random
import sys
def uppercase_and_scramble(line):
ll = list(line)
random.shuffle(ll)
return "".join(ll)
for line in sys.stdin:
print line.upper()What happens if we want to include that function into another file? Then importing do_stuff.py will cause the for loop to run. A much better solution is to do all of the ‘scripty’ stuff in a main function that only gets called if the file is called as a script (as opposed to being imported as a library):
import random
import sys
from optparse import OptionParser
def uppercase_and_scramble(line):
'''
Make the line uppercase, scramble its contents and return the result.
'''
ll = list(line)
random.shuffle(ll)
return "".join(ll)
def main():
usage = """
python do_stuff.py
Process lines from the input.
"""
num_args= 0
parser = OptionParser(usage=usage)
#parser.add_option('-o', '--options', dest='some_option', default='yo', help="Place holder for a real option", type='str')
#parser.add_option('-u', '--useless', dest='uselesss', default=False, action='store_true', help='Another useless option')
(options, args) = parser.parse_args()
if len(args) < num_args:
parser.print_help()
sys.exit(1)
for line in sys.stdin:
print uppercase_and_scramble(line)
if __name__ == '__main__':
main()That’s a lot of code for a simple task. Well it doesn’t necessarily require
much typing to enter thanks to the
SnipMate plugin for
vim. By adding the following code in ~/.vim/snippets/python.snippets we can
create almost all of the code by typing start and hitting tab right
at the beginning of the script.
snippet start
import sys
from optparse import OptionParser
def main():
usage = """
${1:usage}
"""
num_args= 0
parser = OptionParser(usage=usage)
#parser.add_option('-o', '--options', dest='some_option', default='yo', help="Place holder for a real option", type='str')
#parser.add_option('-u', '--useless', dest='uselesss', default=False, action='store_true', help='Another useless option')
(options, args) = parser.parse_args()
if len(args) < num_args:
parser.print_help()
sys.exit(1)
if __name__ == '__main__':
main()This will create the main function and position the cursor within the usage string
thus making it a snap to write some quick documentation of what this script will do.
The num_args variable is there to make sure the user enters the right number of
arguments. Otherwise the script exits with an error. The rest of the processing code
should go directly after the if statement. When scripts are written in this manner,
they can be painlessly turned into libraries at a future point in time.